Yellow rust populations can now be freely browsed on Nextstrain interface
The wheat yellow rust fungus (Puccinia striiformis f.sp. tritici) is the first plant pathogen to be added to an online open source platform, Nextstrain.org. The interactive data visualization tool displays genome data by reconstructing a “phylogeny” that illustrates the differences in the genetic makeup of bacterial, viral and fungal strains. This assists researchers investigating how new pathogen strains emerge and spread. The recent addition of the yellow rust pathogen grants researchers worldwide access to genetic data to study the disease.
When you have a cold, you usually suspect where it came from – maybe even the person who gave it to you. This might make you avoid a certain place, signs of symptoms in another or take extra precautions in future to avoid falling ill again.
In a similar way, when epidemics like Ebola occur, we work out how they spread and use this to inform control strategies to try to prevent the disease from spreading further. By knowing how diseases spread and change over time we can work out how to reduce the likelihood of infection or what control measures to use to stop them spreading. We often think of these strategies with human diseases but we do the same with crops too.
Nextstrain is an online platform to visualise the spread of infectious disease not just over space but also over time. This means you can watch a simulation of how a disease like Zika has evolved and moved over the last 10 years. Genes of interest can be identified and the diversity of the pathogen population explored. This is the same for a number of widely known human bacterial and viral diseases listed on the site.
For the first time however, a fungal plant pathogen can be tracked in the same system.

Wheat rusts and resistance.
Wheat yellow rust is a fungal disease that infects wheat crops around the world. Even with current control measures, yellow rust causes an estimated one billion dollars in lost harvests and control measures each year globally. In fact, a driving part of the agricultural green revolution was Norman Borlaug’s success in breeding rust resistant wheat varieties to feed rising populations. Thanks in part to disease resistant crop varieties, wheat supports a fifth of the calories eaten worldwide and is grown on more of the Earth’s surface than any other crop.
The challenge is that yellow rust, just like other diseases, is constantly changing and some of these changes to its genetic makeup allow it to overcome the resistance of wheat varieties in the field. Today over 80% of the World’s wheat is susceptible to yellow rust disease. Strains of yellow rust also rapidly spread using spores that blow on the wind across fields to infect other susceptible wheat plants and cause great losses to harvest yields. During severe epidemics this can lead to less physical food but also higher prices of staples such as bread that hit the impoverished hardest.
Another part of the problem is that even with today’s technology, breeding new wheat varieties that are resistant to emerging rust strains takes many years. Our current main control to prevent yellow rust from devastating harvests is to apply fungicides to wheat fields. But there is always a risk that the rust pathogens can become resistant to these fungicides and that they are removed from the market due to their potential harm to the wider environment.
With such pressures on these limited methods of rust control, effective protection of global wheat crops depends upon on a greater understanding of the fungal pathogen and how rust strains are spreading through farmers fields.
Nextstrain benefits for researchers
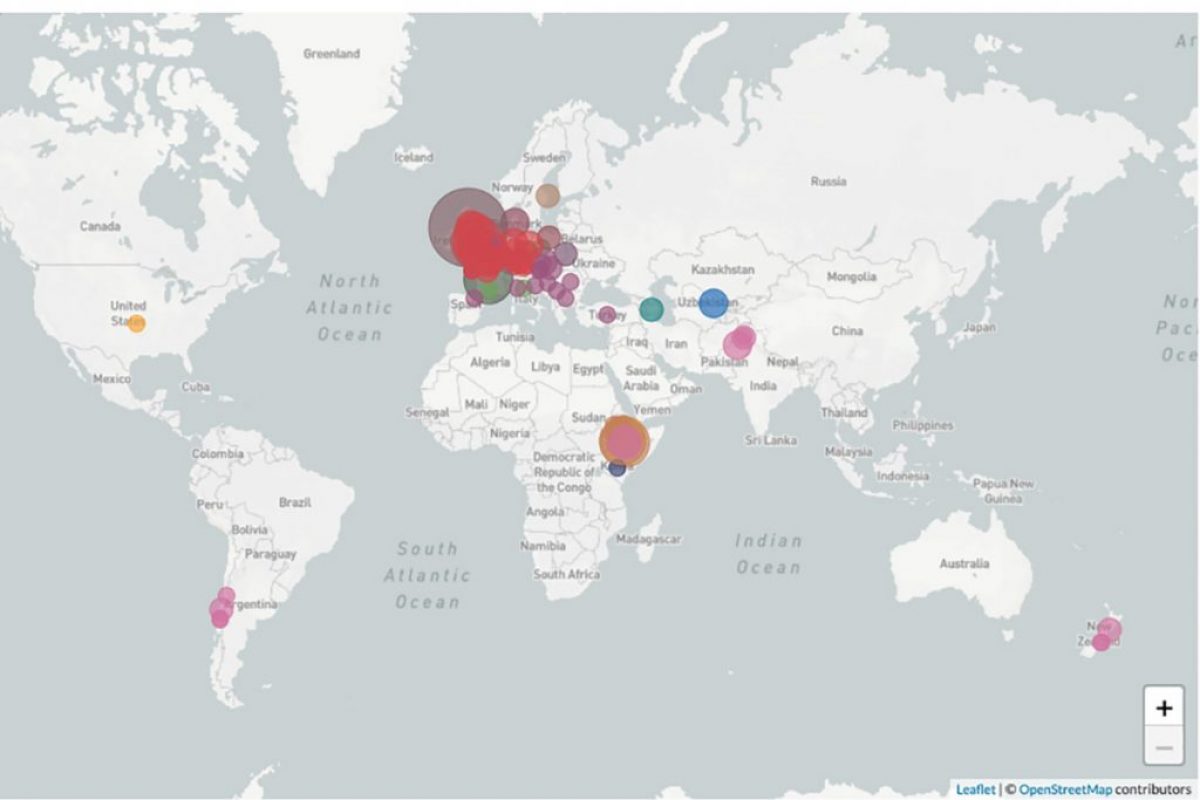
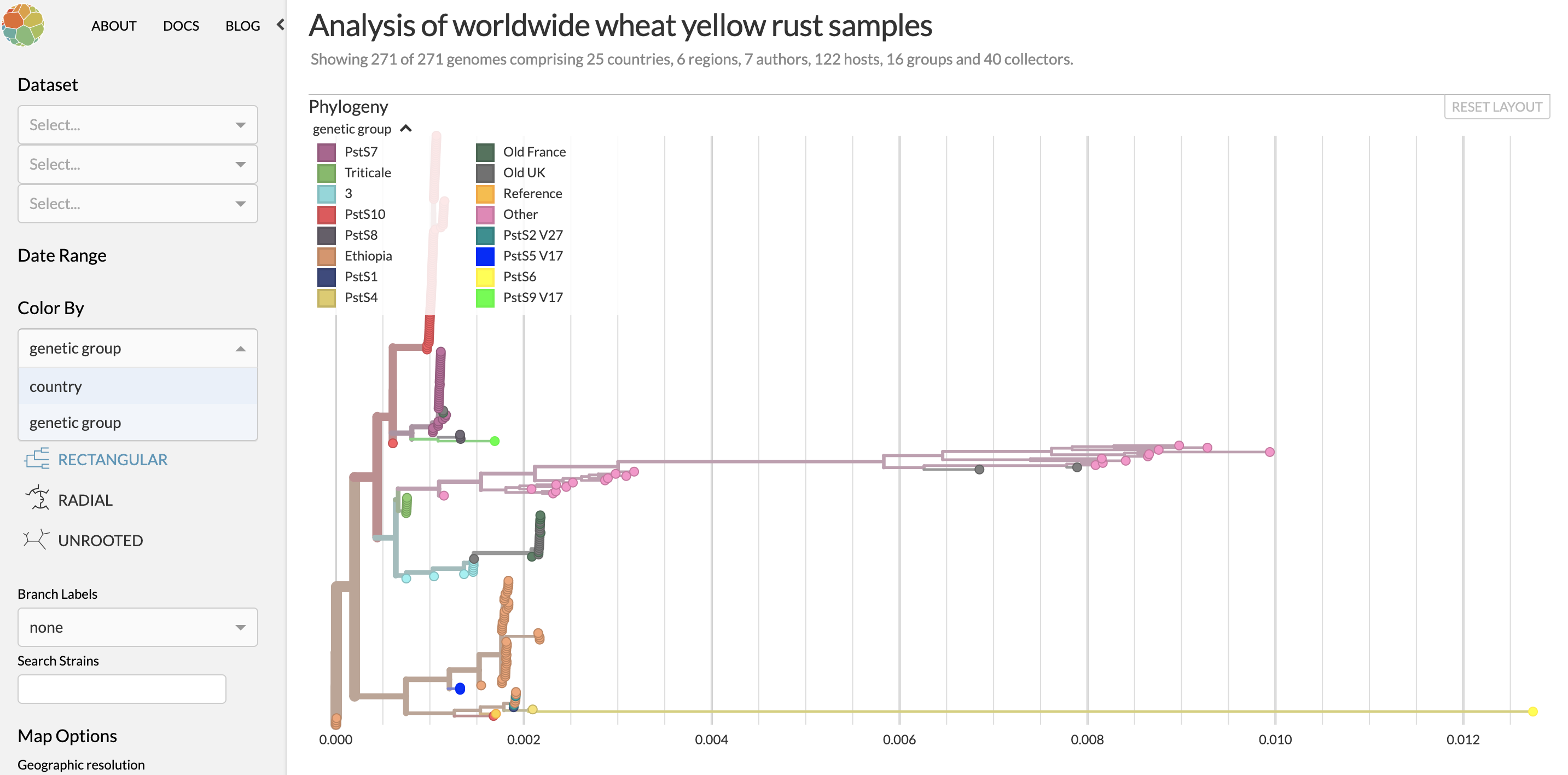
The nextstrain visual display shows a collection of rust strains – separate genetic individuals from the field. Strains that are similar to each other in their genetic makeup are grouped into genetic groups and arranged to show the amount of genetic difference between strains. This is shown as a “phylogeny” that is similar to a family tree. Users can use this interface to explore the rust population and patterns in spread of individual strains to inform future research.
Each rust strain can be easily traced to the research group that found it and even the field the sample was taken from. Geographic distributions can be added to the site in real time meaning that the spread of rust strains across an area can be monitored.
Linking Nextstrain to MARPLE diagnostics
The nextstrain interface is ideally suited to visualizing the small, unified datasets created using our recently developed Mobile and Real-time Plant disease (MARPLE) Diagnostics platform for wheat yellow rust. This diagnostics and surveillance platform generates data on the genetic makeup of individual strains of yellow rust in near real-time. Linking this to the speed of data integration into nextstrain, this provides a swift turn around to show local to regional pathogen movement within the growing season. Publishing the information on platforms such as Nextstrain also means that researchers from labs of any resource size are able to access and interpret the data. This will ultimately help us as a community to better understand how disease outbreaks and new strains spread.