
Project Partners
Diane Saunders
Dave Hodson
Tadessa Daba
Tesfaye Disasa

Rapid Mobile Diagnostics and Surveillance
- Fast: From field sample to strain level identification within 48 hours
- Mobile: Entire lab set up and pipeline can be transported in a single hard case
- Resilient: Pipeline from field to sample can run on inconsistent power and uses reagents that can be kept at room temperature.
- Simple: Easy-to-follow procedure with no prior expertise required.
Complex Fungal pathogens
The MARPLE platform was built initially for wheat yellow rust, which is an obligate biotrophic pathogen that can’t be separated from its wheat host and has a genome > 10,000 larger than for instance the Ebola virus. To address this, we developed a strategy to sequence specific regions of the pathogens genome that can be used to define individual strains and monitor key genes for mutations such as fungicide target genes.


Impact
This new, innovative, data-driven approach provides near real-time data on the rapidly shifting nature of pathogen population structure and dispersal that can feed directly into early-warning systems and control recommendations.

The Pipeline
Sample Collection
Crops showing fungal disease symptoms are identified in the field and small (< 1 cm2) leaf samples collected. These samples are processed using the MARPLE pipeline to confirm the causal pathogen and define the individual strain, whilst also monitoring important genes for mutations such as fungicide target genes.

DNA Extraction
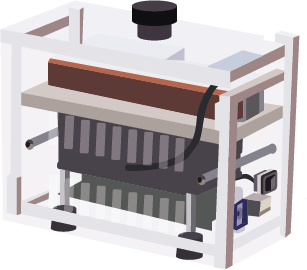
MiniPCR
Mobile PCR amplification: Regions of the pathogens genome that are informative for defining individual strains and target genes of interest are amplified using the polymerase chain reaction (PCR). Currently, a total of 242 genes are PCR amplified across four pools.
Library Preparation and MinION Run

Data Analysis
Access training materials
You can now access the training materials for the MARPLE pipeline. A full guide can now be found below
Funded by








